Using Fish DNA in Threatened Albatross Diets as a Marine Conservation and Management Tool
By Elana Rusnak, SRC Master’s Student
There is an unavoidable interaction between seabirds and the fishing industry, which impacts them through feeding supplementation, resource competition, and incidental mortalities (McInnes et al., 2017). However, resolving these problems is often difficult and requires many resources. Sea-faring birds are attracted to the fish scraps that are discarded from fishing vessels, which oftentimes come from species that are not naturally a part of their diet. Gaining access to this food source may cause an imbalance in food-web structure, allowing gull populations to inflate, or causing albatrosses to prioritize nutritionally-poor food due to its ease of capture (Foster et al., 2017). Moreover, these fisheries may be targeting an important food source for these birds and decreasing their access to it. Understanding the interactions between seabirds and fisheries is necessary for effective ecosystem management.
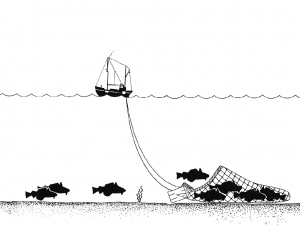
Figure 1: A trawling boat fishing for bottom-dwelling fish to which birds would not normally have access. (Source: NOAA – en:Image:Trawling_Drawing.jpg, Public Domain, https://commons.wikimedia.org/w/index.php?curid=1232501)
In the past, the two main ways to assess seabird diet were looking at their stomach contents, and stable isotope analysis. Unfortunately, neither of these methods yield species-specific results when it comes to what kinds of fish are in these birds’ diets. Recently, a non-invasive process called DNA metabarcoding has been useful in providing high-level specificity in seabird diets when analyzing their waste products. It is also a broad-scale technique, which increases the number of birds and populations that scientists can sample while decreasing the amount of work and time required to do so.

Figure 2: The black-browed albatross (Source: Ed Dunens – Black-browed Albatross, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=64498778)t
The black-browed albatross is found in the southern hemisphere, where its population has been significantly impacted by longline and trawl fisheries. A group of researchers used DNA metabarcoding to assess 6 sites across their breeding range to determine their prey diversity over space and time, identify if any of their prey comes from areas in which there are known fisheries operations, and evaluate potential resource competition or food supplementation by fisheries. Albatross waste was collected, and DNA was extracted from each sample, then cross-referenced with known fish DNA. The researchers found that 91% of their diet consisted of bony fish, with a diversity of 51 species, but the overwhelming majority of birds mostly ate 4 primary species of fish. Samples collected from the 6 breeding sites showed differences in bird diet between sites. A few species of fish identified from the DNA barcoding only live in the northern hemisphere, indicating that these birds are sourcing this prey from fisheries that use those kinds of fish as bait. Depending on the site, between 0-60% of the birds were consuming fishery discards. A few breeding sites were negatively impacted by resource competition, where the fishery was targeting their food source and they therefore did not have access to their normal diets. This study shows that DNA barcoding has provided a means for scientists to prove that improvements in discard management to reduce the number of birds that feed from these vessels would reduce incidental mortality and have major implications for some albatross populations (McInnes et al., 2017).
Works cited
Foster, S., Swann, R. L., & Furness, R. W. (2017). Can changes in fishery landings explain long-term population trends in gulls?. Bird Study, 64(1), 90-97.
McInnes, J. C., Jarman, S. N., Lea, M. A., Raymond, B., Deagle, B. E., Phillips, R. A., … & Gras, M. (2017). DNA metabarcoding as a marine conservation and management tool: a circumpolar examination of fishery discards in the diet of threatened albatrosses. Frontiers in Marine Science, 4, 277.